Protein design
Our protein design team develops and deploys cutting-edge design methods to generate novel proteins for biomedical applications. For evaluation of the designed molecules, we characterize their biophysical properties, determine their structures, and evaluate their activity in vitro, ex vivo and in vivo assays. Our research mainly focuses on the following major areas:
Development of high-performance computational design tools
Despite recent advances in computational protein design, simultaneous enhancements of design throughput and accuracy are continually needed to achieve better success rates at the bench, and tackle more complex design problems. We are continually developing the Damietta design engine; a design framework that leverages the accuracy of established molecular mechanics force fields, while maximizing the throughput of energy calculations through tensorized implementations. While still a bleeding edge technology, we have already successfully used the Damietta engine for different design projects in the lab. Other development domains cover ultra-fast docking and shape similarity algorithms, that enable us to design de novo and rescaffolded binders.
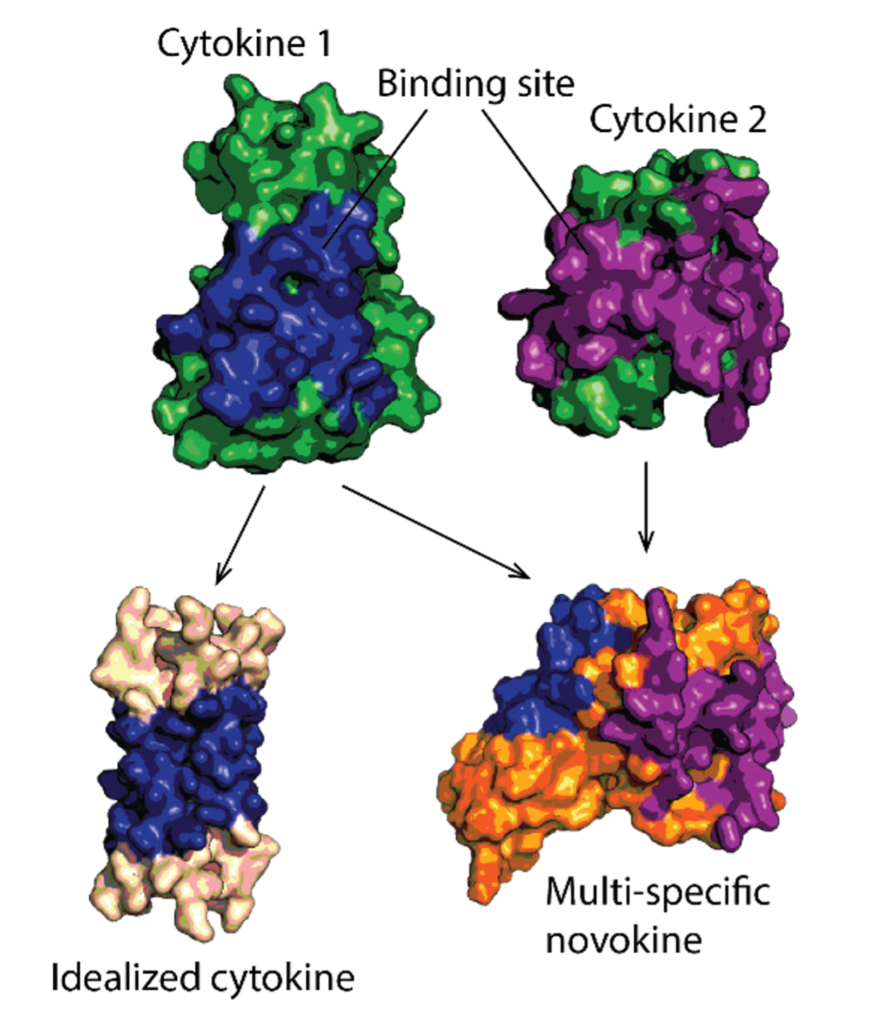
Design of idealised growth factors, cytokines, and novokines
Protein therapeutics must meet a range of requirements to their activity, stability, solubility, aggregation propensity, and production costs. Computational design can offer control over these parameters and greatly facilitate protein drug development. We use a set of in-house developed computational methods for tertiary epitope matching, protein core re-design, interface design, loop design, and accelerated dynamics scoring in order to make idealized therapeutic miniproteins. We design and preclinically-develop idealised growth factors and cytokines with superior properties compared to their natural counterparts. We also use these designs to study the structure-function relationship of receptor modulation. Furthermore, we design novel-function cytokines (i.e. novokines) with the goal of inducing novel signaling patterns in target cells, at a very high targeting specificity.
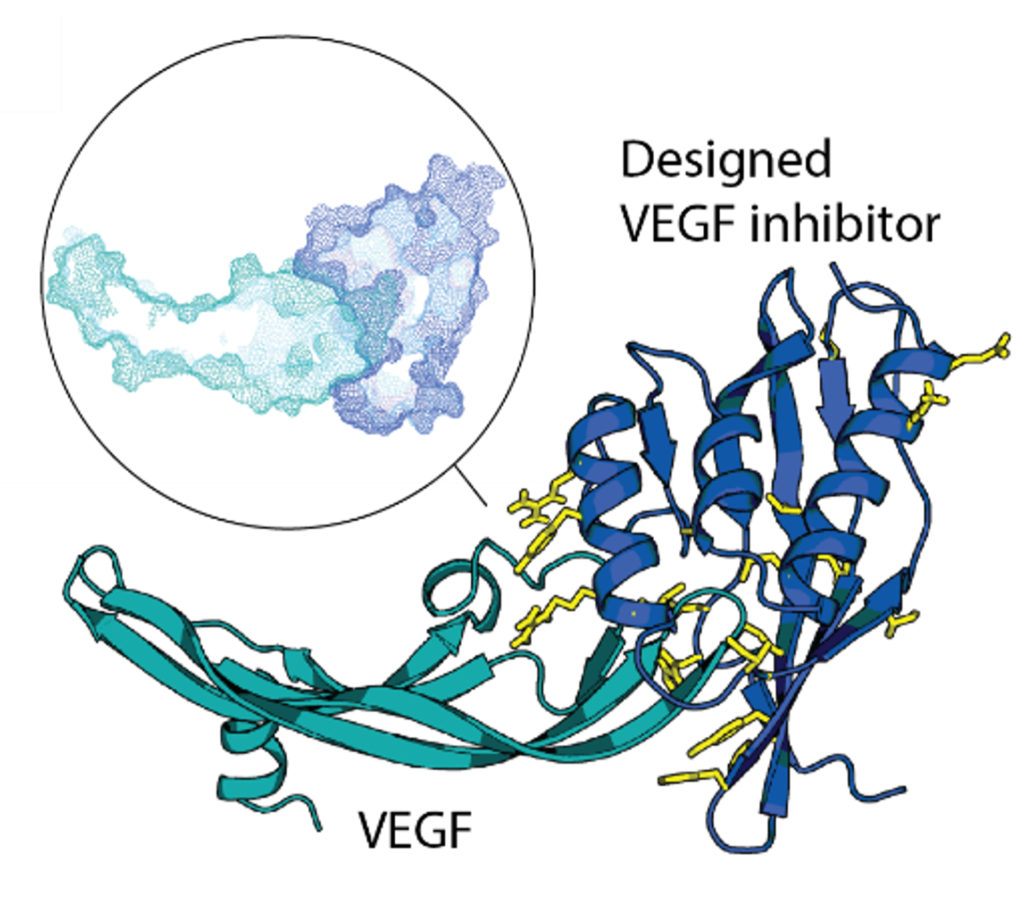
De novo design of epitope-directed binders
De novo design of site-specific binders in a concept-to-hit manner remains a major scientific and technological challenge. We established a novel framework to rapidly create epitope-directed binders using only information on the target structure. Our pipeline takes advantage of a high throughput docking software that evaluates steric complementarity of proteins in a lower dimensional space and enables selection of shape matching scaffolds for further interface design. Previously, we made inhibitors blocking an active site of VEGF, a key angiogenic molecule involved in the pathogenesis of diverse cancers, cardiovascular, and ophthalmic diseases. Moving forward, we work on improvements of the entire pipeline and on design of binders against other relevant targets.
Design of metal-binding proteins
Metalloproteins have a wide range of biomedical applications. For example, they can serve as electron microscopy contrast agents, as emergency antidotes for metal intoxication or as genetically-encodable protein tags for targeted radioimmunotherapy. We aim to design stable, easy-to-purify proteins that would bind copper, lead or other metals with high affinities and high metal binding capacity. For this, we use different approaches, from redesigning natural metalloproteins to mounting multiple metal binding sites onto novel geometrically-accommodating scaffolds.
